| advertisement: compare things at compare-stuff.com! |
We have looked at the single amino acid and duplet patterns in the three
main classes. Tables 3.4 and 3.5 show these
results in detail. Leu, Ala, Glu and Arg are both over-abundant in
mainly-![]() domains and have significant
domains and have significant ![]() at
at ![]() .
Similarly for the mainly-
.
Similarly for the mainly-![]() domains, Thr, Cys, Gly, Ser, Asn, and Val
are found more often than expected. The amino acids favoured by
mixed-
domains, Thr, Cys, Gly, Ser, Asn, and Val
are found more often than expected. The amino acids favoured by
mixed-![]() domains form an intersection between these two sets.
Nakashima et al.nakashima:cpred reported similar trends
in amino acid composition between mainly-
domains form an intersection between these two sets.
Nakashima et al.nakashima:cpred reported similar trends
in amino acid composition between mainly-![]() proteins and
mainly-
proteins and
mainly-![]() proteins. In their work, however, Lys, Met and His are
favoured in
proteins. In their work, however, Lys, Met and His are
favoured in ![]() -proteins, whilst in this statistical study concerning
three classes, they are not found to be important. Differences due to
dataset size and protocol may account for these anomalies, so they are not
dwelled upon.
-proteins, whilst in this statistical study concerning
three classes, they are not found to be important. Differences due to
dataset size and protocol may account for these anomalies, so they are not
dwelled upon.
Comparison with the Chou and Fasmanchou:fasman propensities is
also interesting. Leu, Ala and Glu are the three strongest helix formers
according to Chou and Fasman, whilst Arg, which we find to vary
significantly between the three classes, is `indifferent'. Only Val, Cys
and Thr are Chou-Fasman strand-formers. Ser and Asn are weak
strand-breakers, yet are over-abundant in mainly-![]() proteins in our study.
The apparent paradox here is quickly resolved when one considers that
mainly-
proteins in our study.
The apparent paradox here is quickly resolved when one considers that
mainly-![]() and mainly-
and mainly-![]() proteins are not entirely helix and sheet
respectively; they have turns and loops which connect the secondary
structures. Gly is commonly found in turns and other loops where it allows
specific conformations to be attained (owing to the absence of a
side-chain). In such a role it may be important in mainly-
proteins are not entirely helix and sheet
respectively; they have turns and loops which connect the secondary
structures. Gly is commonly found in turns and other loops where it allows
specific conformations to be attained (owing to the absence of a
side-chain). In such a role it may be important in mainly-![]() proteins.
More discussion of this hypothesis will follow.
proteins.
More discussion of this hypothesis will follow.
| Observed - Expected ( |
||||
| pattern | Mainly Beta | Alpha Beta | Mainly Alpha | |
| T | 92.5 | 280.6 | -213.0 | -67.5 |
| L | 89.2 | -301.7 | 117.8 | 183.9 |
| A | 68.8 | -286.6 | 193.4 | 93.1 |
| C | 67.1 | 119.5 | -105.2 | -14.2 |
| E | 64.6 | -228.7 | 126.1 | 102.5 |
| G | 61.9 | 176.6 | 33.2 | -209.8 |
| S | 60.7 | 233.7 | -170.0 | -63.7 |
| N | 39.6 | 163.0 | -140.4 | -22.5 |
| V | 34.2 | 84.2 | 75.1 | -159.4 |
| R | 33.8 | -129.3 | 35.2 | 94.0 |
| Observed - Expected ( |
||||||
| pattern | Mainly Beta | Alpha Beta | Mainly Alpha | |||
| L | . . | L | 60.0 | -60.7 | 19.4 | 41.3 |
| C | . . | G | 43.3 | 17.1 | -13.9 | -3.2 |
| A | . . | L | 40.9 | -52.9 | 30.7 | 22.2 |
| K | . . | E | 32.0 | -12.5 | -13.8 | 26.3 |
| E | . . | K | 31.5 | -30.8 | 13.4 | 17.4 |
| A | . . | E | 31.5 | -33.1 | 16.8 | 16.3 |
| L | . . | A | 31.4 | -47.8 | 31.8 | 15.9 |
| A | . . | A | 29.4 | -50.8 | 32.8 | 17.9 |
| T | . . | S | 28.5 | 28.7 | -23.2 | -5.4 |
| C | . . | S | 27.8 | 11.6 | -9.1 | -2.4 |
| E | . . | R | 26.6 | -21.6 | 2.6 | 18.9 |
Since class predictions using (i,i+3) duplet composition perform better
than those using amino acid composition, it is not surprising that their
compositional differences (see Table 3.5) are also
interesting. Pairs of helix-preferring residues predominate in both the
helix-containing classes, as might be expected: Leu-X-X-Leu, Ala-X-X-Leu,
Ala-X-X-Glu, Leu-X-X-Ala and Ala-X-X-Ala. However, Lys-X-X-Glu and
Glu-X-X-Lys also exhibit significant ![]() across the three classes.
Lys is only a weak helix former on the Chou-Fasman scale. Intriguingly,
the Lys-X-X-Glu duplet is over-abundant only in mainly-
across the three classes.
Lys is only a weak helix former on the Chou-Fasman scale. Intriguingly,
the Lys-X-X-Glu duplet is over-abundant only in mainly-![]() domains while
the Glu-X-X-Lys duplet is over-abundant in both helix-containing classes.
This will be discussed further below. Mainly-
domains while
the Glu-X-X-Lys duplet is over-abundant in both helix-containing classes.
This will be discussed further below. Mainly-![]() domains possess more
Cys-X-X-Gly, Thr-X-X-Ser and Cys-X-X-Ser pairs.
domains possess more
Cys-X-X-Gly, Thr-X-X-Ser and Cys-X-X-Ser pairs.
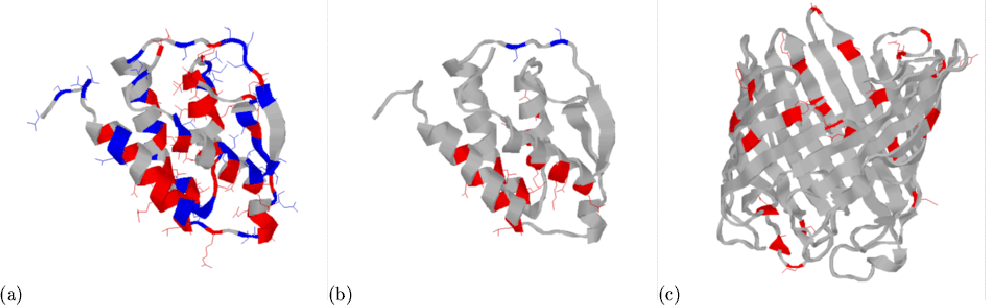 |
We have attempted to give structural explanations for these observations,
by highlighting occurrences of these patterns on 3D rendered structures
(Figure 3.4). Unfortunately, the large number of
structures has prohibited a comprehensive visual examination of the data.
In Figure 3.4(a), the over- and under-abundant amino acids
for mainly-![]() domains (from Table 3.4) are shown in red
and blue respectively on the structure of granulocyte-macrophage
colony-stimulating factor[Diederichs et al.,
1991], domain 1gmfA0 in CATH. In part
(b) of the figure, the same is shown for (i,i+3) duplets (from
Table 3.5). The over-abundant duplets are clearly located in
helices. The clearer distinction (more red than blue) in (b) compared with
(a) is expected since duplets contain much more local structural
information. An inspection of structures containing numerous Lys-X-X-Glu
and Glu-X-X-Lys pairs showed that the majority of these do not form salt
bridges, even though most are situated on the solvent accessible face of
helices where their side-chains can be in close proximity.
domains (from Table 3.4) are shown in red
and blue respectively on the structure of granulocyte-macrophage
colony-stimulating factor[Diederichs et al.,
1991], domain 1gmfA0 in CATH. In part
(b) of the figure, the same is shown for (i,i+3) duplets (from
Table 3.5). The over-abundant duplets are clearly located in
helices. The clearer distinction (more red than blue) in (b) compared with
(a) is expected since duplets contain much more local structural
information. An inspection of structures containing numerous Lys-X-X-Glu
and Glu-X-X-Lys pairs showed that the majority of these do not form salt
bridges, even though most are situated on the solvent accessible face of
helices where their side-chains can be in close proximity.
![]() analysis on the domains used in
Table 3.5 showed many (i,i+3) patterns with structural
preferences: the strongest preferences are for Pro- or Gly-containing
patterns in loop or turn structures (data not shown). The patterns with
class preferences (see Table 3.5) gave lower
analysis on the domains used in
Table 3.5 showed many (i,i+3) patterns with structural
preferences: the strongest preferences are for Pro- or Gly-containing
patterns in loop or turn structures (data not shown). The patterns with
class preferences (see Table 3.5) gave lower ![]() scores,
but these were still significant (
scores,
but these were still significant (![]() ). These results confirm that the
`helix' patterns discussed above (Leu-X-X-Leu, Ala-X-X-Leu, etc.) are
over-abundant in helix.
). These results confirm that the
`helix' patterns discussed above (Leu-X-X-Leu, Ala-X-X-Leu, etc.) are
over-abundant in helix.
The patterns with preferences for mainly-![]() domains are more interesting.
Cys-X-X-Gly has significant (
domains are more interesting.
Cys-X-X-Gly has significant (![]() ) structural preferences and is
over-abundant in turns. Cys-X-X-Ser is also over-abundant in turns, but its
overall distribution is not significant (
) structural preferences and is
over-abundant in turns. Cys-X-X-Ser is also over-abundant in turns, but its
overall distribution is not significant (![]() ). Thr-X-X-Ser is
over-abundant in loops; its overall distribution is not very significant
(
). Thr-X-X-Ser is
over-abundant in loops; its overall distribution is not very significant
(![]() ,
, ![]() ). The interesting observation here is that these
patterns are not strand specific, although many patterns do have strand
preferences (for example Gly-X-X-Ile and Val-X-X-Val). The majority of the
information used in the three class prediction is helix-related (which
explains the better quality predictions for mainly-
). The interesting observation here is that these
patterns are not strand specific, although many patterns do have strand
preferences (for example Gly-X-X-Ile and Val-X-X-Val). The majority of the
information used in the three class prediction is helix-related (which
explains the better quality predictions for mainly-![]() domains).
domains).