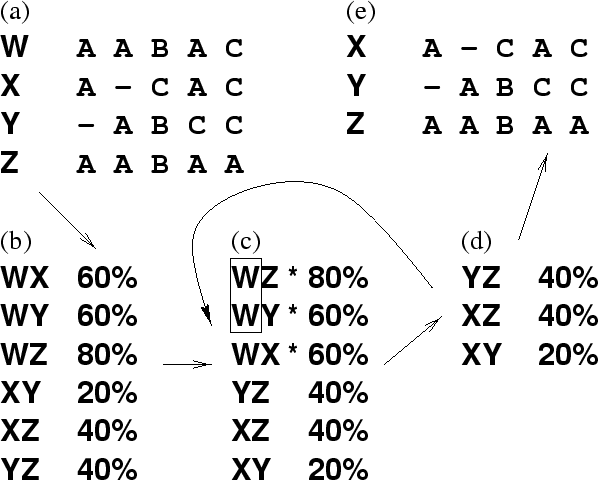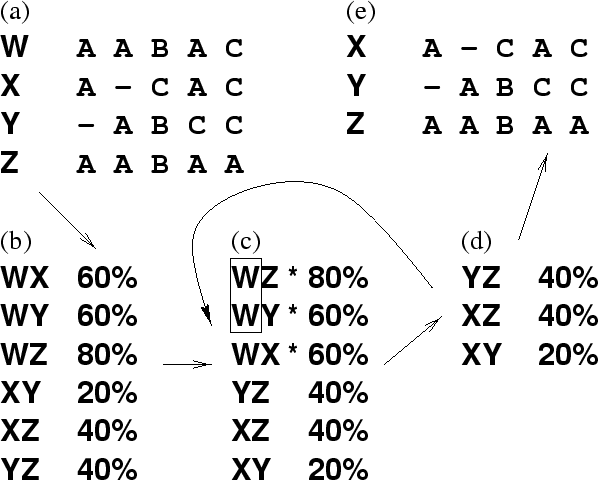Next: CATH descriptions
Up: Gathering multiple sequences for
Previous: Indel removal
Contents
The process by which sequences in a multiple sequence alignment were made
non-redundant on the basis of their pairwise sequence identity is described
in the legend to Figure A.2.
Figure A.2:
Weeding multiple sequences to create a
non-redundant set. The result of this process is a set of sequences with
no more than 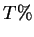 pairwise sequence identity. In this example,
pairwise sequence identity. In this example, 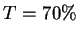 .
See the referring chapters for the exact values used. (a) The multiple
sequence alignment, from which percentage pairwise identities (
.
See the referring chapters for the exact values used. (a) The multiple
sequence alignment, from which percentage pairwise identities (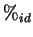 )
(not counting aligned gaps) are calculated directly, as shown in (b). (c)
The sequence pairs are sorted by
)
(not counting aligned gaps) are calculated directly, as shown in (b). (c)
The sequence pairs are sorted by 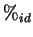 . If the top pair (WZ in the
example) has
. If the top pair (WZ in the
example) has 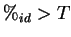 (70%) then the sequence from this pair which next
occurs in the list (W) is eliminated from consideration (i.e. all pairs
containing W are removed, as shown by an asterisk). Step (c) is repeated
until the highest
(70%) then the sequence from this pair which next
occurs in the list (W) is eliminated from consideration (i.e. all pairs
containing W are removed, as shown by an asterisk). Step (c) is repeated
until the highest 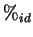 is less than the threshold,
is less than the threshold, 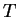 . All sequences
belonging to the remaining pairs (d) form the non-redundant set shown in
(e).
. All sequences
belonging to the remaining pairs (d) form the non-redundant set shown in
(e).
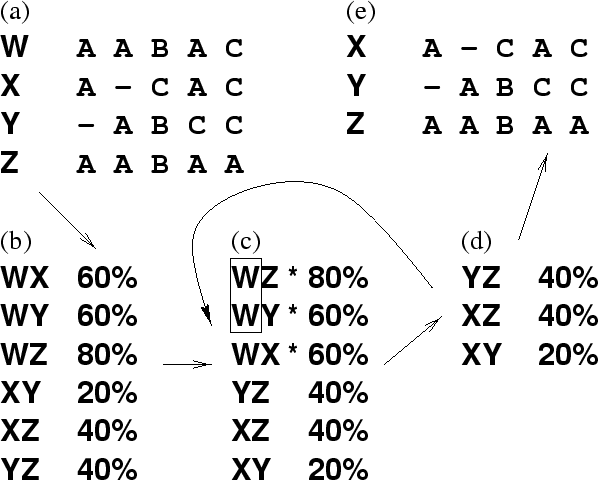 |




Next: CATH descriptions
Up: Gathering multiple sequences for
Previous: Indel removal
Contents
Copyright Bob MacCallum
- DISCLAIMER: this was written in 1997 and may contain out-of-date information.