



Next: Sequence Conservation
Up: Results
Previous: Baseline comparison
Contents
In this section we use the SIVA method to align hydrophobicity profiles,
i.e. the property vector 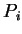 contains as its single component the mean
hydrophobicity,
contains as its single component the mean
hydrophobicity, 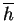 , calculated at sequence position
, calculated at sequence position 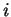 in the
multiple sequence alignment. The summary fold recognition results,
including
in the
multiple sequence alignment. The summary fold recognition results,
including
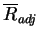 ,
, 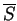 and
and 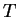 , are presented in
Table 4.4 for the four hydrophobicity scales tested (shown in
Table 4.3). The Kyte and Doolittle hydrophobicity scale
achieves the highest number of correct top-ranking fold recognitions
(
, are presented in
Table 4.4 for the four hydrophobicity scales tested (shown in
Table 4.3). The Kyte and Doolittle hydrophobicity scale
achieves the highest number of correct top-ranking fold recognitions
(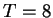 ); a little better than the Smith Waterman results. Amongst the
trials using hydrophobicity related information, the Eisenberg and
McLachlan direction of hydrophobic moment measure gives the best overall
ranking (
); a little better than the Smith Waterman results. Amongst the
trials using hydrophobicity related information, the Eisenberg and
McLachlan direction of hydrophobic moment measure gives the best overall
ranking (
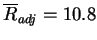 ) and alignments (
) and alignments (
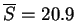 ).
The performance measures vary considerably within the three fold topologies
also shown in Table 4.4. Each of the four measures performs
best in one or other category. Most notably the hydrophobic moment
direction gives the best ranking for the immunoglobulin-like folds
(2.60.40), but gives undistinguished alignments.
).
The performance measures vary considerably within the three fold topologies
also shown in Table 4.4. Each of the four measures performs
best in one or other category. Most notably the hydrophobic moment
direction gives the best ranking for the immunoglobulin-like folds
(2.60.40), but gives undistinguished alignments.
Table 4.3:
Four hydrophobicity related amino acid indices.
| amino acid |
Kyte & |
Bull & |
Radzicka & |
Eisenberg & |
| |
Doolittle1 |
Breese2 |
Wolfenden3 |
McLachlan4 |
| I |
4.5 |
-2.26 |
1.31 |
0.99 |
| V |
4.2 |
-1.56 |
1.09 |
0.84 |
| L |
3.8 |
-2.46 |
1.21 |
0.89 |
| F |
2.8 |
-2.33 |
1.27 |
0.92 |
| C |
2.5 |
-0.45 |
1.36 |
0.76 |
| M |
1.9 |
-1.47 |
1.27 |
0.94 |
| A |
1.8 |
-0.20 |
-0.06 |
0 (n/a)5 |
| G |
-0.4 |
0.00 |
-0.41 |
0 (n/a) |
| T |
-0.7 |
-0.52 |
-0.27 |
0.09 |
| S |
-0.8 |
-0.39 |
-0.50 |
-0.67 |
| W |
-0.9 |
-2.01 |
0.88 |
0.67 |
| Y |
-1.3 |
-2.24 |
0.33 |
-0.93 |
| P |
-1.6 |
-0.98 |
0.00 |
0.22 |
| H |
-3.2 |
-0.12 |
0.49 |
-0.75 |
| Q |
-3.5 |
0.16 |
-0.73 |
-1.00 |
| N |
-3.5 |
0.08 |
-0.48 |
-0.86 |
| E |
-3.5 |
-0.30 |
-0.77 |
-0.89 |
| D |
-3.5 |
-0.20 |
-0.80 |
-0.98 |
| K |
-3.9 |
-0.35 |
-1.18 |
-0.99 |
| R |
-4.5 |
-0.12 |
-0.84 |
-0.96 |
|
- 1 [Kyte & Doolittle, 1982]
- 2 [Bull & Breese, 1974]
- 3 [Radzicka & Wolfenden, 1988]
- 4 [Eisenberg & McLachlan, 1986]
- 5 Too few side-chain atoms to calculate moment
|
Table 4.4:
Summary of fold recognition using alignment of raw sequence derived information.
| Sequence |
whole library |
3.40.3301 |
3.20.40 |
2.60.40 |
| information |
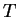 |
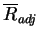 |
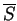 |
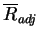 |
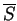 |
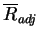 |
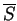 |
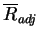 |
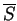 |
| Kyte & Doolittle |
8 |
11.4 |
22.4 |
6.7 |
18.1 |
5.4 |
26.0 |
12.9 |
30.1 |
| |
|
|
|
|
|
|
|
|
|
| Radzicka & |
6 |
12.0 |
22.6 |
5.1 |
17.3 |
6.2 |
32.5 |
21.2 |
28.5 |
| Wolfenden |
|
|
|
|
|
|
|
|
|
| Bull & Breese |
7 |
14.2 |
23.2 |
5.6 |
20.5 |
7.8 |
29.7 |
16.0 |
27.1 |
| |
|
|
|
|
|
|
|
|
|
| Eisenberg & |
6 |
10.8 |
20.9 |
7.0 |
16.8 |
5.7 |
29.8 |
9.2 |
29.2 |
| McLachlan |
|
|
|
|
|
|
|
|
|
| Conservation |
2 |
17.0 |
25.1 |
10.9 |
22.8 |
9.7 |
31.3 |
11.7 |
19.3 |
| |
|
|
|
|
|
|
|
|
|
| Conserved |
9 |
10.8 |
23.2 |
5.7 |
19.9 |
6.2 |
27.1 |
17.9 |
29.5 |
| Hydrophobicity |
|
|
|
|
|
|
|
|
|
| Hydrophobicity & |
10 |
10.5 |
23.6 |
7.5 |
21.2 |
4.6 |
22.5 |
13.4 |
30.3 |
| Conservation 20:1 |
|
|
|
|
|
|
|
|
|
| DSC prediction |
8 |
9.0 |
26.8 |
5.2 |
22.3 |
5.3 |
33.3 |
3.7 |
25.9 |
| |
|
|
|
|
|
|
|
|
|
| Hydrophobicity & |
10 |
9.5 |
21.5 |
5.4 |
17.6 |
4.1 |
23.4 |
6.3 |
29.0 |
| DSC prediction 2:1 |
|
|
|
|
|
|
|
|
|
| Hydrophobicity & |
13 |
7.7 |
22.2 |
4.4 |
18.0 |
3.6 |
23.9 |
2.2 |
29.1 |
| DSC prediction 1:1 |
|
|
|
|
|
|
|
|
|
| Hydrophobicity & |
10 |
7.6 |
24.7 |
5.2 |
20.9 |
4.1 |
30.8 |
2.0 |
28.2 |
| DSC prediction 1:2 |
|
|
|
|
|
|
|
|
|
|
- 1 see Appendix B for
descriptions of CATH codes
|
Table 4.5:
Summary of fold recognition results
using alignments of hydrophobicity (Kyte and Doolittle). Only
`correct' query-library pairs are shown.
| rank by |
|
domain |
CATH |
length |
|
| all1 |
query2 |
Z-score |
query |
library |
topology |
query |
library |
 3 3 |
| 1 |
1 |
1.940 |
5p2100 |
1hurA0 |
3.40.330 |
166 |
180 |
1.0 |
| 2 |
1 |
1.860 |
1hurA0 |
5p2100 |
3.40.330 |
180 |
166 |
0.9 |
| 5 |
1 |
0.904 |
1pii01 |
1tpfA0 |
3.20.40 |
261 |
250 |
22.0 |
| 15 |
1 |
0.775 |
1tpfA0 |
1pii01 |
3.20.40 |
250 |
261 |
25.4 |
| 17 |
1 |
0.758 |
1dgd02 |
1aam02 |
3.40.640 |
264 |
271 |
17.8 |
| 26 |
1 |
0.696 |
1llo00 |
1pii01 |
3.20.40 |
273 |
261 |
24.4 |
| 28 |
3 |
0.684 |
1atnA2 |
1atr03 |
3.30.420 |
108 |
107 |
4.2 |
| 32 |
2 |
0.673 |
1atr03 |
1atnA2 |
3.30.420 |
107 |
108 |
3.9 |
| 35 |
1 |
0.655 |
1ntr00 |
4fxn00 |
3.40.330 |
124 |
138 |
5.0 |
| 47 |
3 |
0.608 |
1llo00 |
1tpfA0 |
3.20.40 |
273 |
250 |
9.4 |
| 48 |
1 |
0.603 |
4fxn00 |
1ntr00 |
3.40.330 |
138 |
124 |
3.7 |
|
- 1 global ranking (out of 2214 query-library pairs) by Z-score calculated per query
- 2 ranked by alignment score for each query (out of 82
pairwise alignments) -- therefore a `1' in this column indicates a correct
top ranking alignment.
- 3 mean alignment shift
|
Table 4.6:
Summary fold recognition
results using alignments of hydrophobicity (Bull and Breese). Only
`correct' query-library pairs are shown.
| rank by |
|
domain |
CATH |
length |
|
| all |
query |
Z-score |
query |
library |
topology |
query |
library |
 |
| 1 |
1 |
2.466 |
1hurA0 |
5p2100 |
3.40.330 |
180 |
166 |
0.8 |
| 2 |
1 |
2.335 |
5p2100 |
1hurA0 |
3.40.330 |
166 |
180 |
0.7 |
| 7 |
3 |
0.982 |
1atr03 |
1atnA2 |
3.30.420 |
107 |
108 |
2.6 |
| 11 |
1 |
0.828 |
1atnA2 |
1atr03 |
3.30.420 |
108 |
107 |
4.3 |
| 19 |
3 |
0.761 |
1cnd01 |
1pkm03 |
2.40.90 |
106 |
103 |
18.3 |
| 27 |
3 |
0.724 |
1llo00 |
1nal10 |
3.20.40 |
273 |
291 |
21.8 |
| 33 |
1 |
0.701 |
1pkm03 |
1cnd01 |
2.40.90 |
103 |
106 |
14.1 |
| 41 |
4 |
0.674 |
1dgd02 |
1aam02 |
3.40.640 |
264 |
271 |
19.4 |
| 49 |
5 |
0.642 |
1nal10 |
1llo00 |
3.20.40 |
291 |
273 |
21.0 |
| 51 |
1 |
0.641 |
1ntr00 |
5p2100 |
3.40.330 |
124 |
166 |
3.9 |
| 58 |
1 |
0.613 |
4fxn00 |
1ntr00 |
3.40.330 |
138 |
124 |
2.0 |
| 60 |
8 |
0.607 |
1aam02 |
1dgd02 |
3.40.640 |
271 |
264 |
23.2 |
| 64 |
5 |
0.600 |
1llo00 |
1tpfA0 |
3.20.40 |
273 |
250 |
10.2 |
| 71 |
7 |
0.584 |
1llo00 |
1pii01 |
3.20.40 |
273 |
261 |
37.2 |
| 72 |
2 |
0.574 |
1ntr00 |
4fxn00 |
3.40.330 |
124 |
138 |
2.5 |
| 73 |
1 |
0.569 |
2ohxA2 |
1pnrA3 |
3.40.330 |
139 |
147 |
8.9 |
|
Table 4.7:
Summary of fold recognition results using
alignments of hydrophobic moment direction (Eisenberg and McLachlan). Only
`correct' query-library pairs are shown.
| rank by |
|
domain |
CATH |
length |
|
| all |
query |
Z-score |
query |
library |
topology |
query |
library |
 |
| 1 |
1 |
2.085 |
5p2100 |
1hurA0 |
3.40.330 |
166 |
180 |
0.6 |
| 2 |
1 |
1.980 |
1hurA0 |
5p2100 |
3.40.330 |
180 |
166 |
0.5 |
| 8 |
1 |
0.813 |
1dgd02 |
1aam02 |
3.40.640 |
264 |
271 |
15.7 |
| 10 |
1 |
0.781 |
1ntr00 |
4fxn00 |
3.40.330 |
124 |
138 |
4.0 |
| 16 |
1 |
0.718 |
1pii01 |
1tpfA0 |
3.20.40 |
261 |
250 |
22.2 |
| 33 |
4 |
0.643 |
1pii01 |
1nal10 |
3.20.40 |
261 |
291 |
9.3 |
| 34 |
1 |
0.640 |
4fxn00 |
1ntr00 |
3.40.330 |
138 |
124 |
2.8 |
| 38 |
2 |
0.623 |
1llo00 |
1nal10 |
3.20.40 |
273 |
291 |
21.8 |
| 44 |
2 |
0.606 |
1nal10 |
1pii01 |
3.20.40 |
291 |
261 |
8.9 |
| 45 |
3 |
0.602 |
1nal10 |
1tpfA0 |
3.20.40 |
291 |
250 |
17.1 |
|
More detailed results for Kyte and Doolittle hydrophobicity, Bull and
Breese hydrophobicity, and the hydrophobic moment direction are given in
Tables 4.5, 4.6 and 4.7.
These tables show all the correct top-ranking fold predictions, and other
correct pairings. The tables are sorted by Z-score (these are calculated
separately for each query). From these tables one can judge what
proportion of pairings are correct above a certain Z-score threshold. In
these experiments (and almost all that follow), the two domains 5p2100 and
1hurA0 align with each other to give the highest ranking Z-scores. These
domains were also detected by the Smith Waterman method and are clearly the
`easiest' pair to recognise, however the other correct top ranking pairs
are not always the same as the Smith Waterman hits (in
Table 4.2). Note also that the Smith Waterman algorithm has
identified similarities between domains of very different length (for
example 1tpfA0 and 1pii02) through the use of a local alignment algorithm.
In contrast the SIVA top hits all have much smaller differences in length.




Next: Sequence Conservation
Up: Results
Previous: Baseline comparison
Contents
Copyright Bob MacCallum
- DISCLAIMER: this was written in 1997 and may contain out-of-date information.
![]() contains as its single component the mean
hydrophobicity,
contains as its single component the mean
hydrophobicity, ![]() , calculated at sequence position
, calculated at sequence position ![]() in the
multiple sequence alignment. The summary fold recognition results,
including
in the
multiple sequence alignment. The summary fold recognition results,
including
![]() ,
, ![]() and
and ![]() , are presented in
Table 4.4 for the four hydrophobicity scales tested (shown in
Table 4.3). The Kyte and Doolittle hydrophobicity scale
achieves the highest number of correct top-ranking fold recognitions
(
, are presented in
Table 4.4 for the four hydrophobicity scales tested (shown in
Table 4.3). The Kyte and Doolittle hydrophobicity scale
achieves the highest number of correct top-ranking fold recognitions
(![]() ); a little better than the Smith Waterman results. Amongst the
trials using hydrophobicity related information, the Eisenberg and
McLachlan direction of hydrophobic moment measure gives the best overall
ranking (
); a little better than the Smith Waterman results. Amongst the
trials using hydrophobicity related information, the Eisenberg and
McLachlan direction of hydrophobic moment measure gives the best overall
ranking (
![]() ) and alignments (
) and alignments (
![]() ).
The performance measures vary considerably within the three fold topologies
also shown in Table 4.4. Each of the four measures performs
best in one or other category. Most notably the hydrophobic moment
direction gives the best ranking for the immunoglobulin-like folds
(2.60.40), but gives undistinguished alignments.
).
The performance measures vary considerably within the three fold topologies
also shown in Table 4.4. Each of the four measures performs
best in one or other category. Most notably the hydrophobic moment
direction gives the best ranking for the immunoglobulin-like folds
(2.60.40), but gives undistinguished alignments.