| advertisement: compare things at compare-stuff.com! |
So far, this study has paid little attention to the structural relatedness
of the folds in our dataset. Inspection of hit lists shows that some
queries yield a succession of correct top hits, whilst other queries of the
same fold topology yield none. Are similarities being missed for these
queries because they are too weak, or because the method fails to
discriminate signal from noise for these examples? In
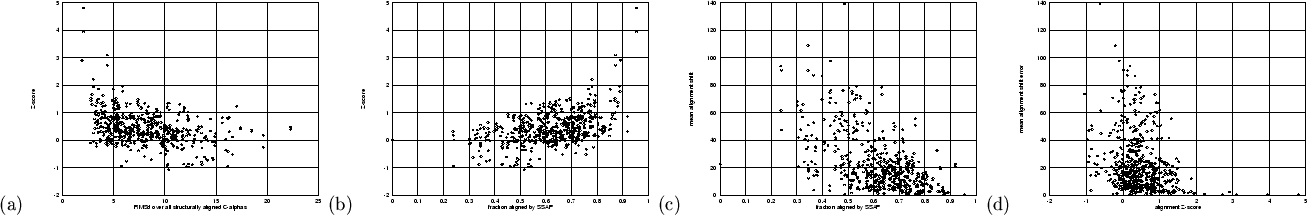
Figure 4.4 the relationships between structural similarity
and fold recognition performance are explored. Figure 4.4(a)
shows that a greater number of similar folds (with lower RMSd) are more
likely to be aligned with a higher Z-score. However, the majority of
similar pairs, with RMSd![]() Å for example, have been aligned unreliably
(
Å for example, have been aligned unreliably
(![]() ). These are pairs of folds which the method has missed.
Interestingly, the inverse situation also exists: reasonably high scoring
alignments (
). These are pairs of folds which the method has missed.
Interestingly, the inverse situation also exists: reasonably high scoring
alignments (![]() ) are made for quite dissimilar pairs (RMSd
) are made for quite dissimilar pairs (RMSd![]() Å),
indicating that close structural similarity is not necessarily required in
order for fold recognition to be successful. Figure 4.4(b)
shows basically the same trend, this time using the fraction of aligned
structure rather than RMSd, as a semi-independent measure of similarity.
We cannot yet explain why so many pairs are missed, and will undertake an
analysis of their alignments and look in more detail at the structural
comparisons.
Å),
indicating that close structural similarity is not necessarily required in
order for fold recognition to be successful. Figure 4.4(b)
shows basically the same trend, this time using the fraction of aligned
structure rather than RMSd, as a semi-independent measure of similarity.
We cannot yet explain why so many pairs are missed, and will undertake an
analysis of their alignments and look in more detail at the structural
comparisons.
 |