| advertisement: compare things at compare-stuff.com! |
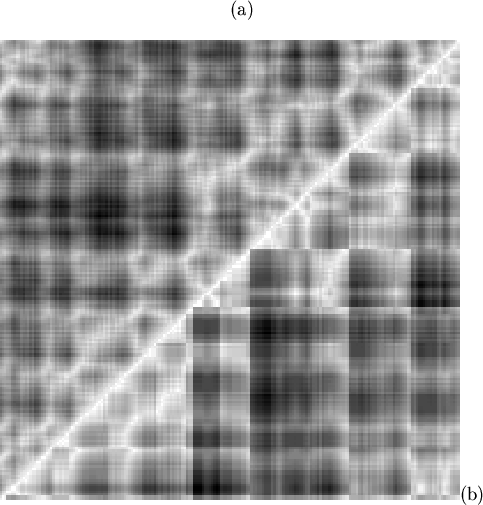
A number of sharp edges can be seen in the Figure 5.4(b);
these correspond to discontinuities in the mapping (in
Figure 5.3, and see explanation in
Figure 5.2). All but one of the six discontinuities with a
map distance greater than 5 occur at the carboxy-termini of the strand
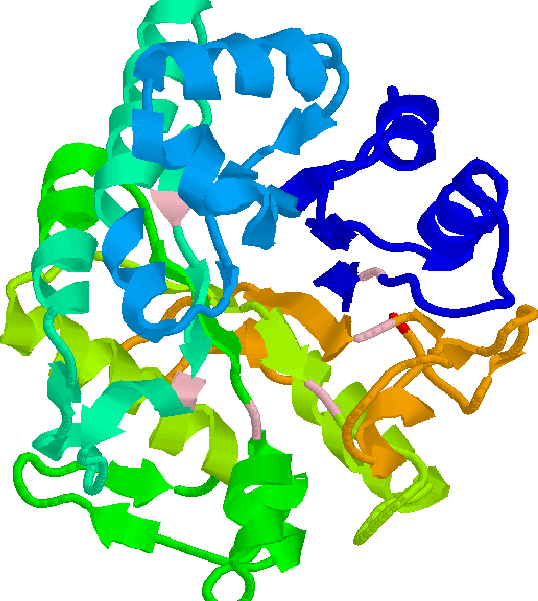
residues, shown in pink in Figure 5.5. The majority
of TIM-barrel proteins are enzymes, and the active site is always found at
the carboxy-end of the ![]() -strands. Structural studies[Urfer & Kirschner, 1992, and
references therein] suggest that the helix-loop-strand units
(at the amino-terminus of the barrel) are more conserved and important for
stabilisation of the fold, than the strand-loop-helix units at the opposite
(active-site) end of the barrel. It is interesting that the
discontinuities in the mapping of 1ghsA0 are at the less-stable end of the
-strands. Structural studies[Urfer & Kirschner, 1992, and
references therein] suggest that the helix-loop-strand units
(at the amino-terminus of the barrel) are more conserved and important for
stabilisation of the fold, than the strand-loop-helix units at the opposite
(active-site) end of the barrel. It is interesting that the
discontinuities in the mapping of 1ghsA0 are at the less-stable end of the
![]() -strands, and that the more structurally important residues are not
interrupted by discontinuities. Examination of seven other TIM-barrel
domains (1pii01, 1amg01, 1llo00, 1ads00, 1tpfA0, 1nal10, and 1xyzA0) chosen
objectively showed that this observation was part of a general
trend
-strands, and that the more structurally important residues are not
interrupted by discontinuities. Examination of seven other TIM-barrel
domains (1pii01, 1amg01, 1llo00, 1ads00, 1tpfA0, 1nal10, and 1xyzA0) chosen
objectively showed that this observation was part of a general
trend![]() . Discontinuities may
be forced to occur where fewer (stabilising) inter-residue contacts are
made. The residues identified by this method may be suitable points for
the insertion of motifs or domains in the engineering of proteins with
novel functions.
. Discontinuities may
be forced to occur where fewer (stabilising) inter-residue contacts are
made. The residues identified by this method may be suitable points for
the insertion of motifs or domains in the engineering of proteins with
novel functions.
 |
 |